Advanced coarse-grained model for fast simulation of nascent polypeptide chain dynamics within the ribosome
We present an advanced coarse-grained model for simulating the dynamics of nascent polypeptide chains within the ribosome
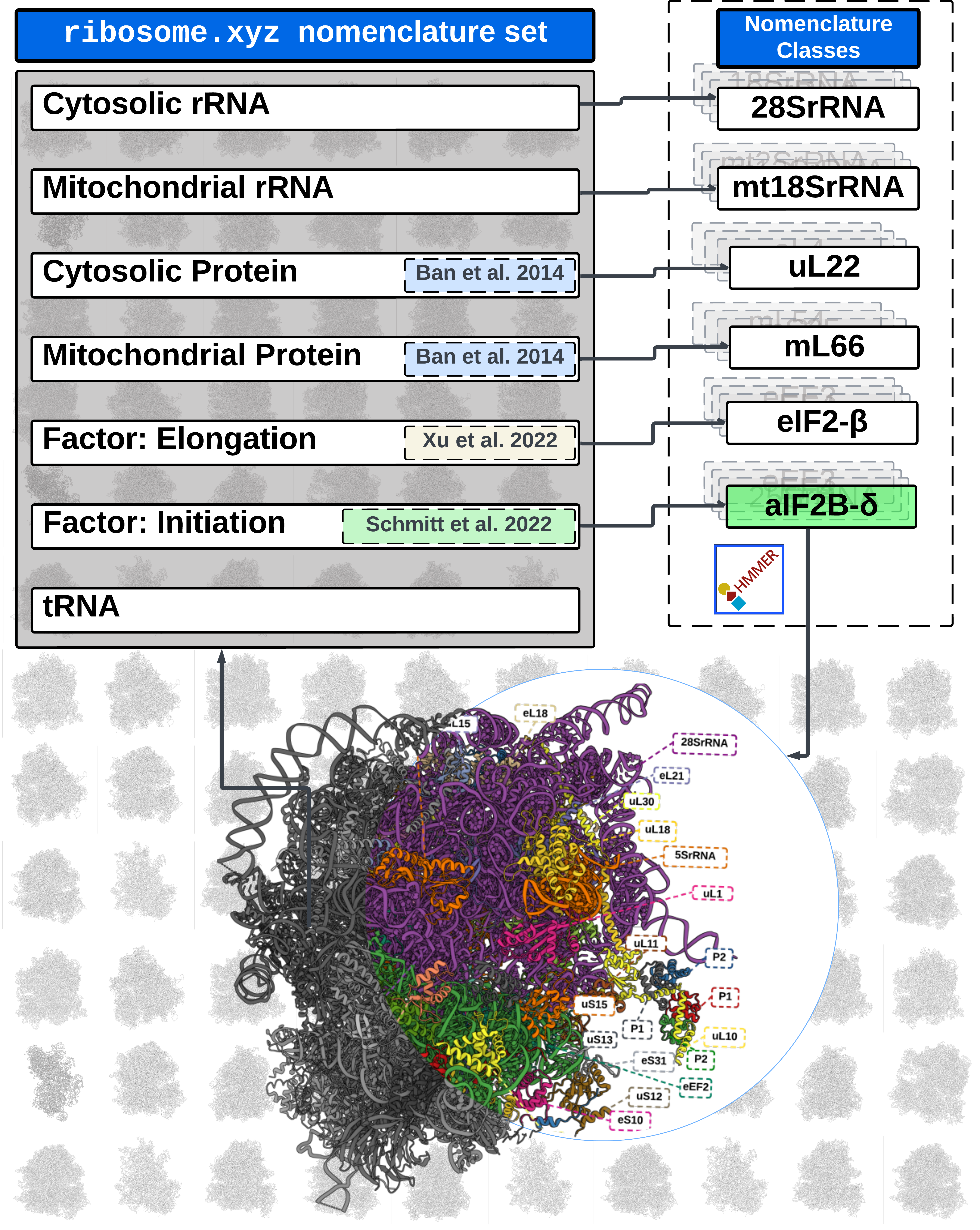
RiboXYZ: a comprehensive database for visualizing and analyzing ribosome structures
RiboXYZ is a comprehensive database designed for the visualization and analysis of ribosome structures from various organisms and functional states. Visit the project at ribosome.xyz.
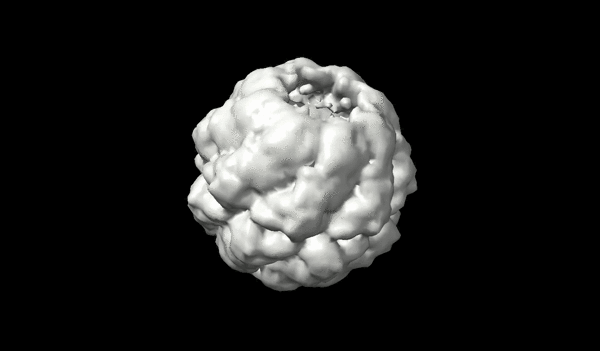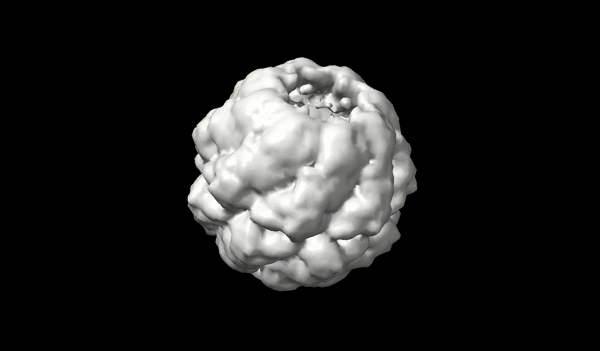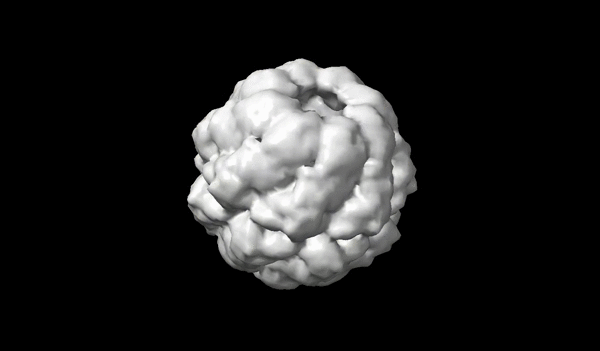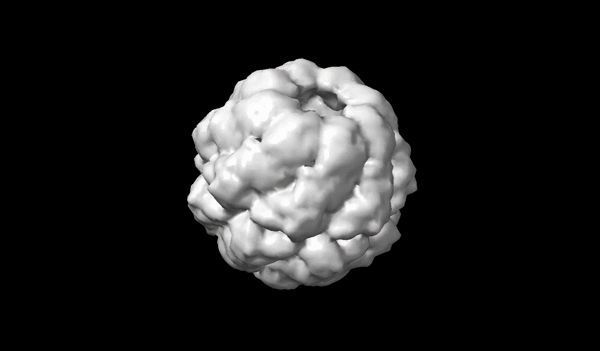
Application of transport-based metric for continuous interpolation between cryo-EM density maps
We present an approach to interpolating between cryo-EM density maps based on the theory of optimal transport, allowing for a more physically meaningful representation of conformational changes.
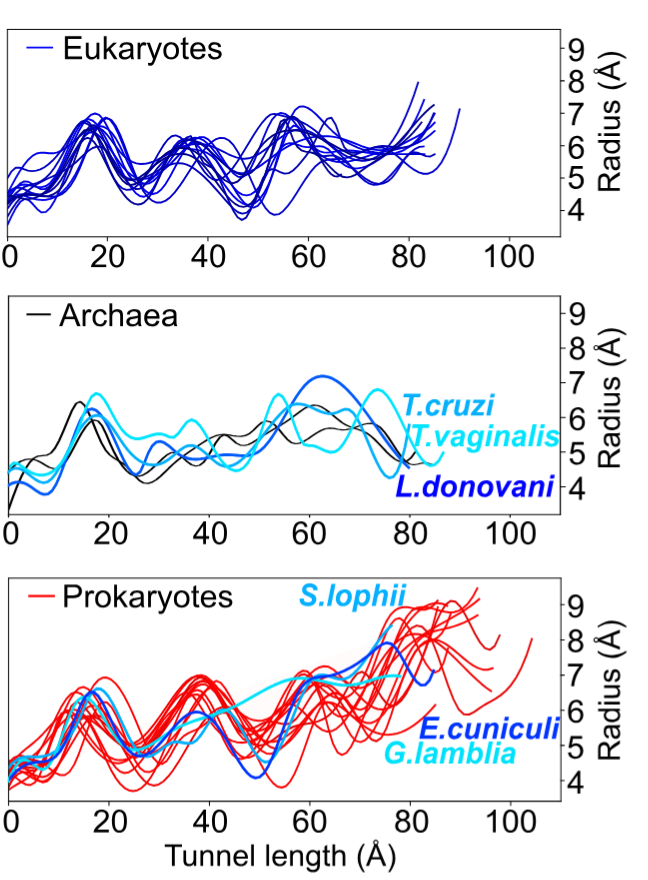
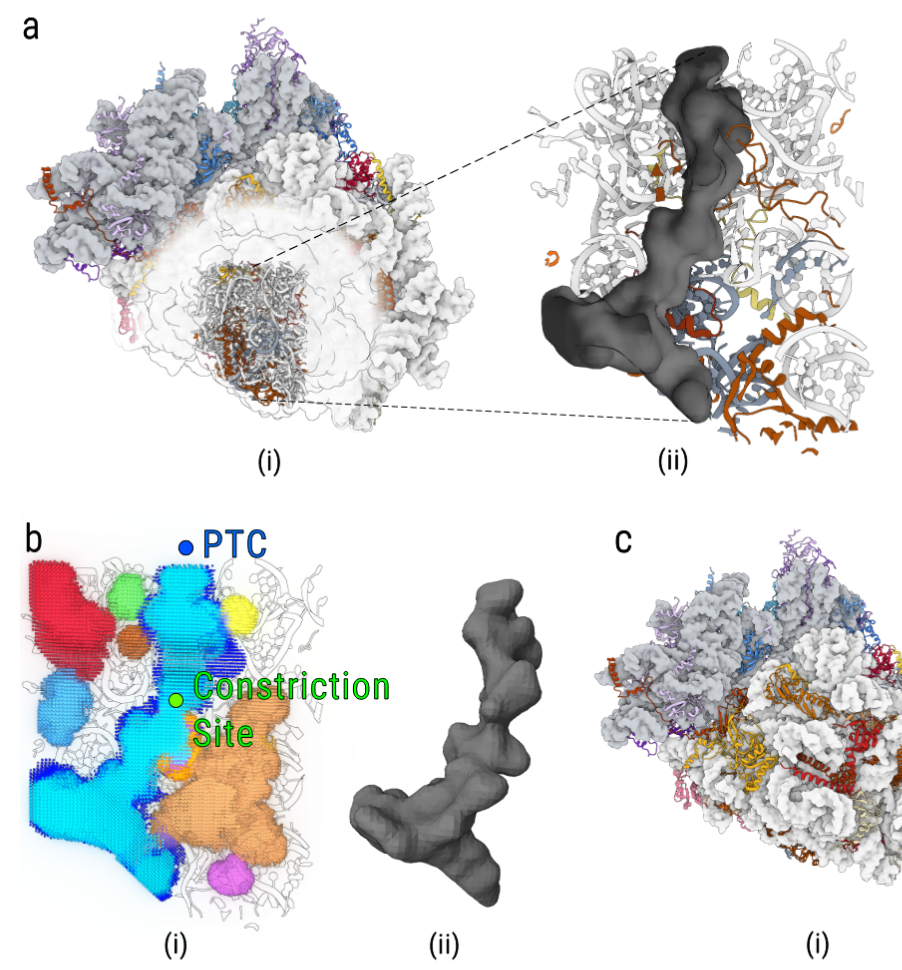
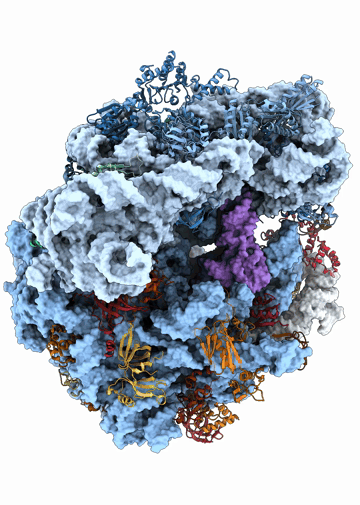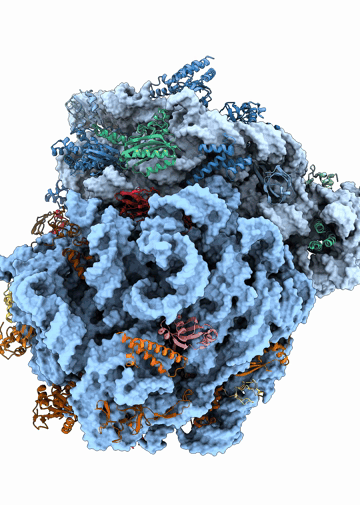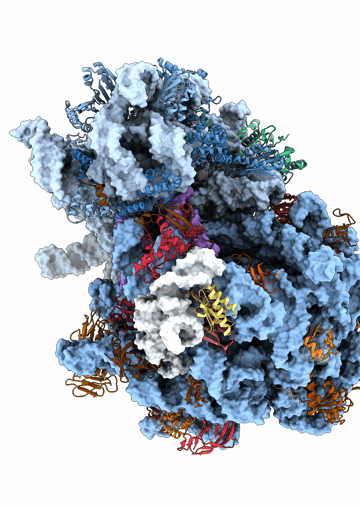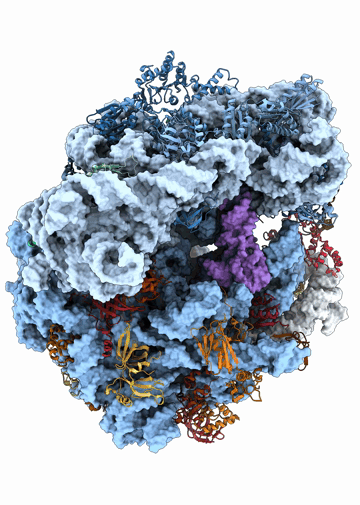
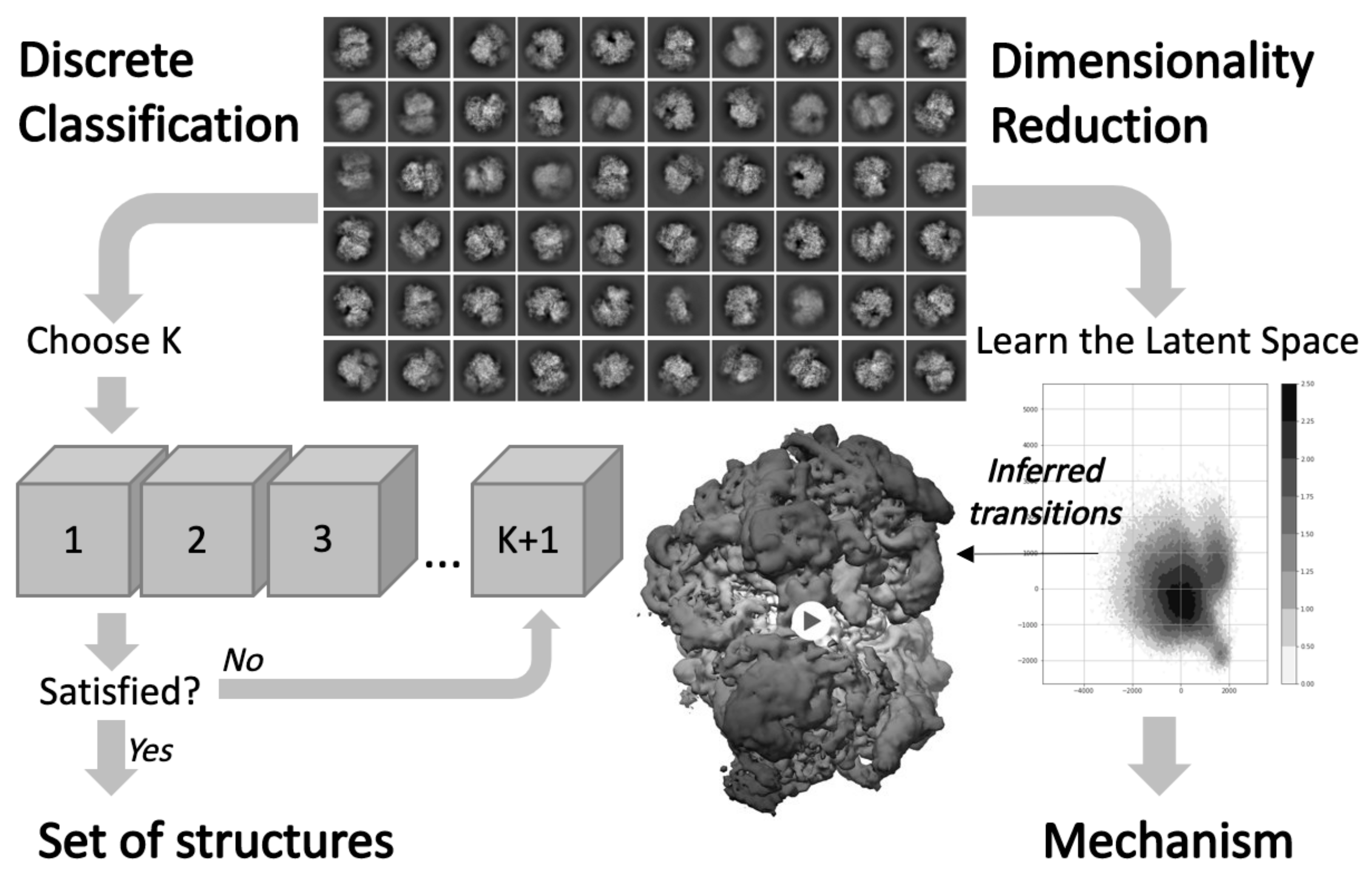
Structural heterogeneities of the ribosome: new frontiers and opportunities for cryo-EM
This paper explores the structural heterogeneities of the ribosome across evolutionary, cellular, temporal and conformational landscapes, highlighting new frontiers for cryo-EM.
Computational methods for unraveling ribosome structural heterogeneity
We shared some computational methods for analyzing ribosome structural heterogeneity.